This web page was produced as an assignment for Gen677 at UW-Madison Spring 2009
NFATc
Nucleotide Sequence:
The nucleotide sequence for NFATc was obtained from the NCBI website. The sequence in humans is found to be 76117153 base pairs in length and can also be know as: NFAT2, NF-ATC, MGC138448, NFATC1. The NFATc coding sequence resides on the 18th chromosome. There are five know isoforms for NFATc and the full sequence to Isoform B is represented through this sequence link. Isoform B will be the primary sequence used for further analysis of this gene.
DNA Motifs
Using MOTIF with a cut of score of 85, 15 motifs were found for the nucleotide sequence of Isoform B. One of the motifs uncovered was of IGF-Binding origin. It was found at 15 different positions in the sequence. IGF-Binding is a signature of proteins involved in the binding of insulin-like growth factors. This makes sense as NFATc has been shown to be involved in the reuglation of insulin homeostasis. (1) A cysteine-rich domain signature of integrin beta chain was identified as being located at 114 different locations in the seguence. The high level of integrin beta chains indicates that this sequence may belong to a transmembrane protein, as integrins a generally involved in the binding of ligands in the extracellular matrix. (2) Whether or not this is true of the NFATc protein will require a look into the gene ontology of the sequence. An interesting motif that was only found at one site in the sequence was one of J-ACTX family signature. This is unusual in that this motif belongs to a characteristic janus-faced atracotoxin which is an insect-specific excitatory neurotoxin. (3) This just goes to show that these alogrithms do not necessarily give a complete and accurate look into the motifs specific to a protein.
NFATc Isoforms
Homology Between Isoforms:
T-coffee was used to align the five isoforms of NFATc. The alignment can be seen by downloading the PDF file (below left). When compared, the isoforms show a relative similarity of 87 which is high, and generally expected for isoforms in the same organism. As seen in the output from T-coffee, the sequences show a strong homology beginning at approximately the 300th coding sequence and continuing until approximately the 700th. The only largely striking difference between the isoforms is seen in Isoform D which is much shorter in sequence length the others. However, where sequence is available for Isoform D it shows very strong homologys
Homology Between Organisms
T-coffee was then used to establish a homology between NFATc in five different organisms: homo sapiens (human), mus musculus (mouse), danio rerio (zebra muscles), gallus gallus (chicken), and bos taurus (cow). The gene retains the same title in all of the organisms with only minor technical changes. In mus musculus it is Nfatc1 and in danio rerio nfatc1 (Information obtained from Homologene). The comparative homology can be seen by downloading the PDF below or clicking on this link. The overall homology is scored as 79, which is not as good as when human isoforms were compared. The homology for the five organisms is strongest in the center portion of the sequence and worst near the c-terminus of the protein. The large area of highly conserved sequence suggests that it may contain the active domain of the protein structure that may conserve function across organisms.
| Comparative Homology | |
| File Size: | 72 kb |
| File Type: | |
Phylogeny
ISOFORM PHYLOGENY
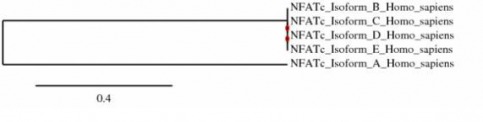
Phylogeny.fr was used to construct a tree between the five isoforms of NFATc in humans. The figure above shows the tree that was generated. It can be seen that Isoforms B-E are all very closely related, and Isoform A forms an outgroup. The fact the Isoform A is shown as the most unrelated is interesting in the fact that when comparing the isoforms using T-coffee, Isoform D had the lowest homology rating of 82, compared to the rest which had ratings between 86-89. (See the Isoform Homology pdf file under NFATc Isoforms)
PHYLOGENY BETWEEN ORGANISMS
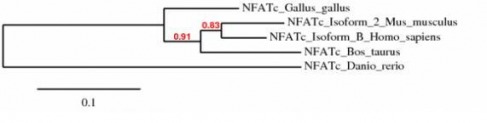
Phylogeny.fr was used again here to compile a tree for the five homologs. As shown above, the human and mouse homologs are the most closely related. The cow and the chicken are also among those closest related while the zebra mussel appears to form an outgroup.
References:
1. Yang et al.(2006) Role of Transcription Factor NFAT in Glucose and Insulin Homeostasis. MCB 2006 October; 26(20). Retrieved February 16, 2009 from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1636854
2. Humphries et al.(2006) Integrin ligands at a glance. Journal of Cell Science, 119.Retrieved February 16, 2009 from http://jcs.biologists.org/cgi/content/full/119/19/3901
3. Maggio et al.(2002) Scanning mutagenesis of a Janus-faced atracotoxin reveals a bipartite suface patch that is essential for neurotoxic function. JBC. Retrieved February 16, 2009 from http://www.jbc.org/cgi/content/abstract/M202297200v1
Algorithmic Websites:
NCBI: http://www.ncbi.nlm.nih.gov/
MOTIF: http://motif.genome.jp/
T-coffee: http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=tcoffee
Homologene: http://www.ncbi.nlm.nih.gov/homologene
Phylogeny.fr: http://www.phylogeny.fr/
*All the figures shown were generated using these algorithms
Margaret Noll, [email protected], last updated 4/26/2009, http://www.gen677.weebly.com
